
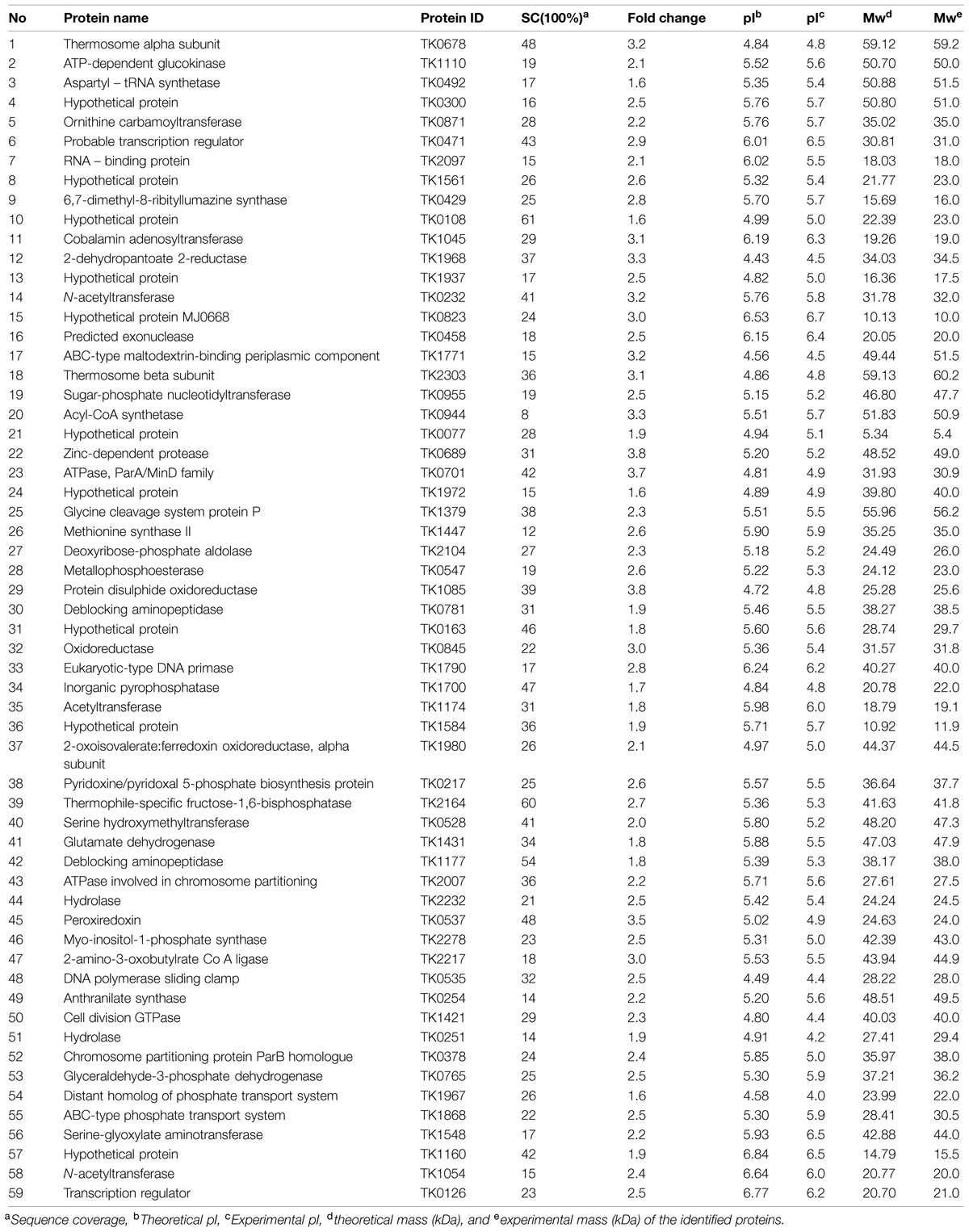
A variety of modeling environments have been developed to simulate biochemical reactions and gene transcription kinetics ( Endy and Brent 2001), cellular physiology ( Loew and Schaff 2001), and metabolic control ( Mendes 1997). Several case studies of Cytoscape plug-ins are surveyed, including a search for interaction pathways correlating with changes in gene expression, a study of protein complexes involved in cellular recovery to DNA damage, inference of a combined physical/functional interaction network for Halobacterium, and an interface to detailed stochastic/kinetic gene regulatory models.Ĭomputer-aided models of biological networks are a cornerstone of systems biology. The Core is extensible through a straightforward plug-in architecture, allowing rapid development of additional computational analyses and features.
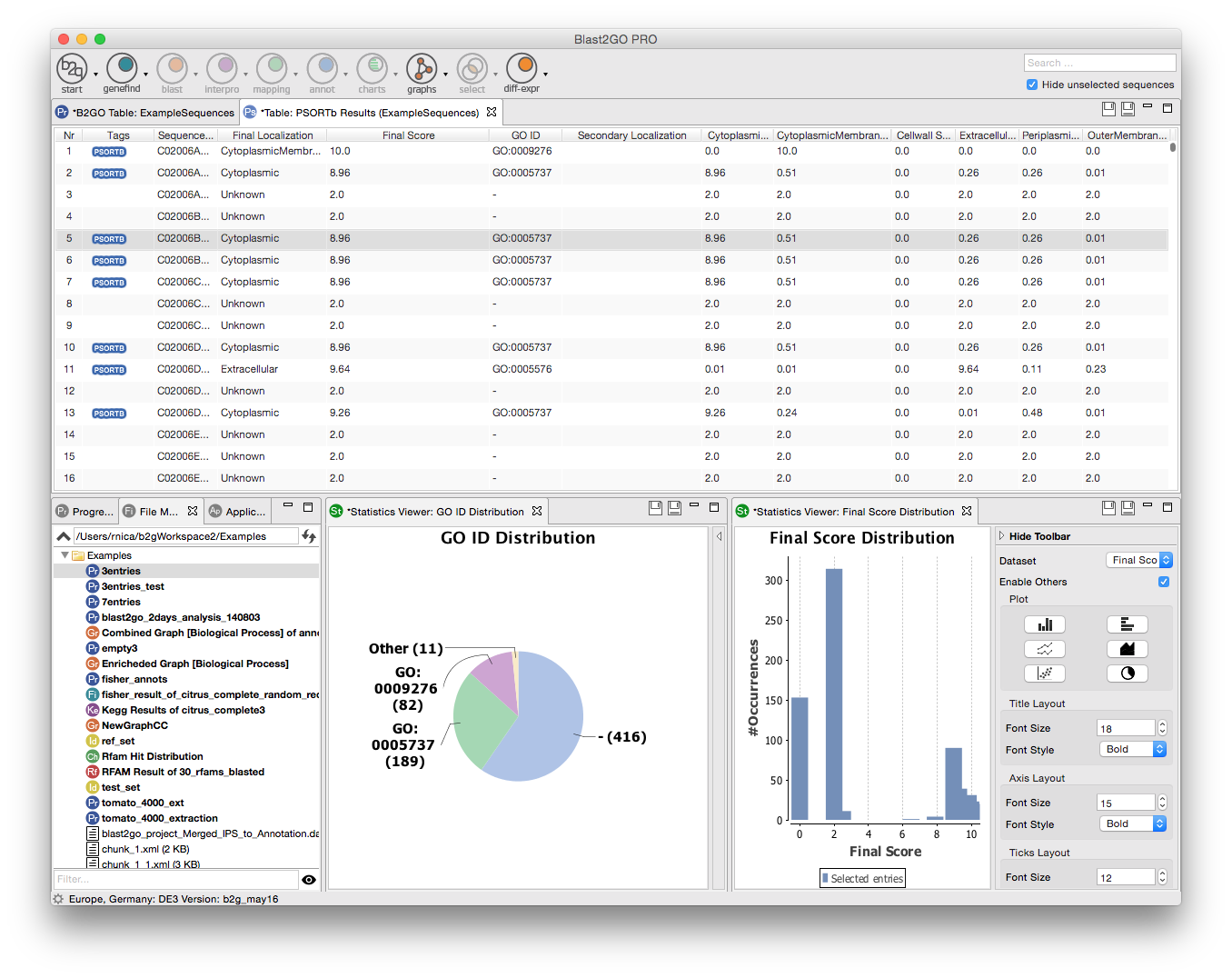
Cytoscape and blast2go software#
Cytoscape's software Core provides basic functionality to layout and query the network to visually integrate the network with expression profiles, phenotypes, and other molecular states and to link the network to databases of functional annotations. Although applicable to any system of molecular components and interactions, Cytoscape is most powerful when used in conjunction with large databases of protein-protein, protein-DNA, and genetic interactions that are increasingly available for humans and model organisms.
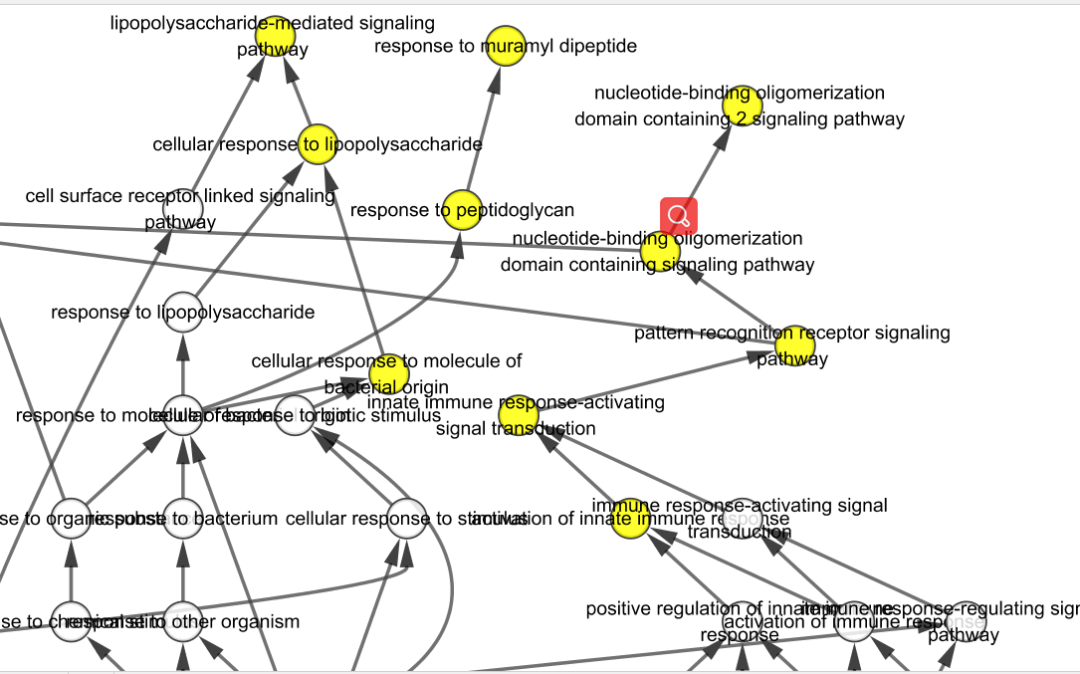
Gene products listed in either the study or population set files that are not present in the annotation file will be ignored.Cytoscape is an open source software project for integrating biomolecular interaction networks with high-throughput expression data and other molecular states into a unified conceptual framework. Dashed edges indicate that one or more intermediate terms were ommited from the graph. In addition to the name of each GO term, the graph
The graph is colored by p-value: terms with p-value above cut-off appear in white and the color gets darker as the p-value decreases The graph file can be either a png image, an svg image, or a text file for importing into cytoscape (together with the result file).The result file is a tabular list of all GO terms present in the study set and their respective p-values.GOEnrichment produces a tabular result file and a graph file for each GO type (MF - Molecular Function, BP - Biological Process and CC - Cellular Component): Optionally, a list of gene products comprising the population set (if none is submitted, the population set will be the set of gene products listed in the annotation file).A list of gene products comprising the study set (a flat text file with one gene product per line).from BioMart) with gene product ids in the first column and GO terms in the second one. A tabular annotation file in GAF ( ) format, BLAST2GO format, or a simple two-column table (e.g.A Gene Ontology file in either OBO or OWL format (see ).GOEnrichment is a Java application that can be used to analyze gene product sets (e.g., from microarray or RNAseq experiments) for enriched GO terms.


 0 kommentar(er)
0 kommentar(er)
